Structure-activity relationship for the folding intermediate-selective inhibition of DYRK1A.
Miyazaki, Y., Kikuchi, M., Umezawa, K., Descamps, A., Nakamura, D., Furuie, G., Sumida, T., Saito, K., Kimura, N., Niwa, T., Sumida, Y., Umehara, T., Hosoya, T., Kii, I.(2022) Eur J Med Chem 227: 113948-113948
- PubMed: 34742017
- DOI: https://doi.org/10.1016/j.ejmech.2021.113948
- Primary Citation of Related Structures:
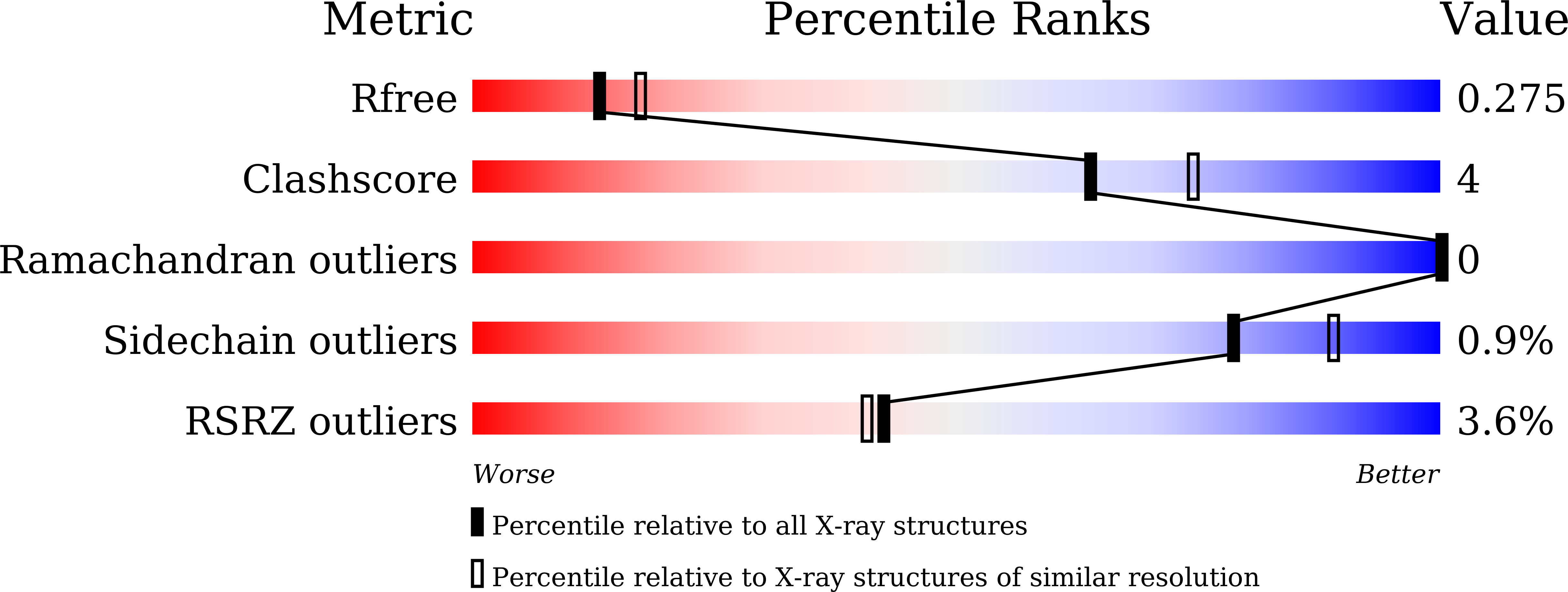
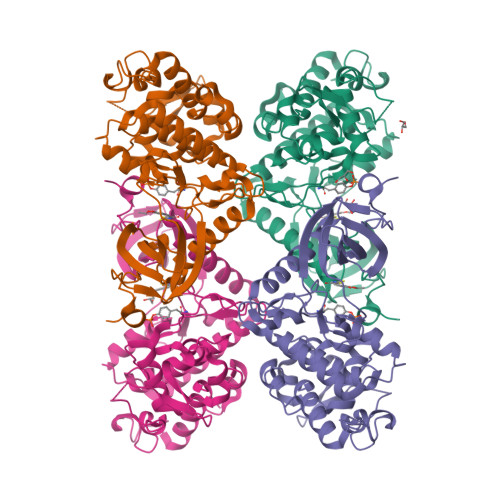
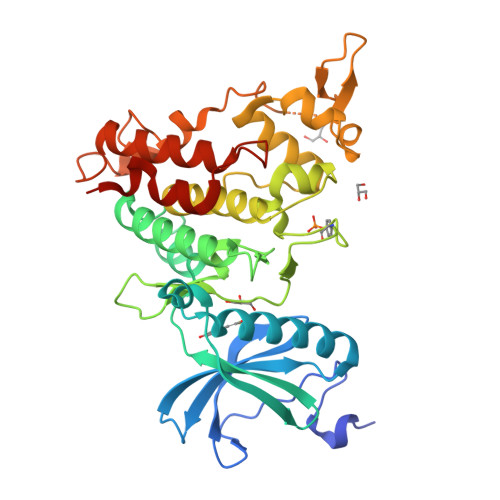
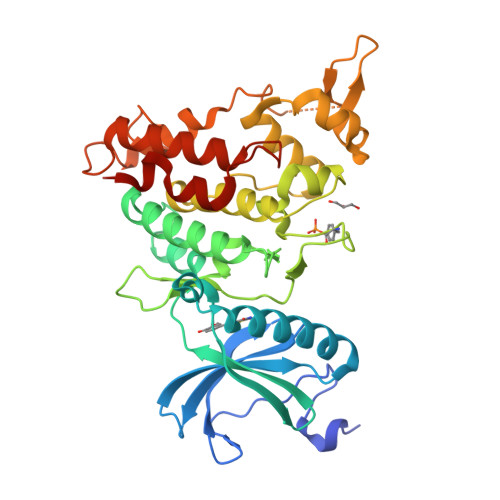
7FHS, 7FHT - PubMed Abstract:
DYRK1A phosphorylates proteins involved in neurological disorders in an intermolecular manner. Meanwhile, during the protein folding process of DYRK1A, a transitional folding intermediate catalyzes the intramolecular autophosphorylation required for the "one-off" inceptive activation and stabilization. In our previous study, a small molecule termed FINDY (1) was identified, which inhibits the folding intermediate-catalyzed intramolecular autophosphorylation of DYRK1A but not the folded state-catalyzed intermolecular phosphorylation. However, the structural features of FINDY (1) responsible for this intermediate-selective inhibition remain elusive. In this study, structural derivatives of FINDY (1) were designed and synthesized according to its predicted binding mode in the ATP pocket of DYRK1A. Quantitative structure-activity relationship (QSAR) of the derivatives revealed that the selectivity against the folding intermediate is determined by steric hindrance between the bulky hydrophobic moiety of the derivatives and the entrance to the pocket. In addition, a potent derivative 3 was identified, which inhibited the folding intermediate more strongly than FINDY (1); it was designated as dp-FINDY. Although dp-FINDY (3) did not inhibit the folded state, as well as FINDY (1), it inhibited the intramolecular autophosphorylation of DYRK1A in an in vitro cell-free protein synthesis assay. Furthermore, dp-FINDY (3) destabilized endogenous DYRK1A in HEK293 cells. This study provides structural insights into the folding intermediate-selective inhibition of DYRK1A and expands the chemical options for the design of a kinase inhibitor.
Organizational Affiliation:
Laboratory for Drug Target Research, Department of Agriculture, Graduate School of Science and Technology, Shinshu University, 8304 Minami-Minowa, Kami-Ina, Nagano, 399-4598, Japan.